Automated Testing
Introduction
Being able to demonstrate that a process generates the right results is
important in any field of research, whether it's software generating those
results or not. So when writing software we need to ask ourselves some key
questions:
- Does the code we develop work the way it should do?
- Can we (and others) verify these assertions for themselves?
- Perhaps most importantly, to what extent are we confident of the accuracy of results that software produces?
If we are unable to demonstrate that our software fulfills these criteria, why
would anyone use it? Having well-defined tests for our software is useful for
this, but manually testing software can prove an expensive process.
Automation can help, and automation where possible is a good thing - it enables
us to define a potentially complex process in a repeatable way that is far less
prone to error than manual approaches. Once defined, automation can also save us
a lot of effort, particularly in the long run. In this episode we'll look into
techniques of automated testing to improve the predictability of a software
change, make development more productive, and help us produce code that works as
expected and produces desired results.
What Is Software Testing?
For the sake of argument, if each line we write has a 99% chance of being right,
then a 70-line program will be wrong more than half the time. We need to do
better than that, which means we need to test our software to catch these
mistakes.
We can and should extensively test our software manually, and manual testing is
well-suited to testing aspects such as graphical user interfaces and reconciling
visual outputs against inputs. However, even with a good test plan, manual
testing is very time consuming and prone to error. Another style of testing is
automated testing, where we write code that tests the functions of our software.
Since computers are very good and efficient at automating repetitive tasks, we
should take advantage of this wherever possible.
There are three main types of automated tests:
- Unit tests are tests for fairly small and specific units of functionality, e.g. determining that a particular function returns output as expected given specific inputs.
- Functional or integration tests work at a higher level, and test functional paths through your code, e.g. given some specific inputs, a set of interconnected functions across a number of modules (or the entire code) produce the expected result. These are particularly useful for exposing faults in how functional units interact.
- Regression tests make sure that your program's output hasn't changed, for example after making changes your code to add new functionality or fix a bug.
For the purposes of this course, we'll focus on unit tests. But the principles
and practices we'll talk about can be built on and applied to the other types of
tests too.
Inflammation Data Analysis
We will be using a simple inflammation data analysis python package to demonstrate the use of automated testing. Let's download this now. First, create a new directory inflammation and
cd to it:mkdir inflammation cd inflammation
If on WSL or Linux (e.g. Ubuntu or the Ubuntu VM), then do:
wget https://train.oxrse.uk/material/HPCu/software_architecture_and_design/procedural/inflammation/inflammation.zip
Or, if on a Mac, do:
curl -LO https://train.oxrse.uk/material/HPCu/software_architecture_and_design/procedural/inflammation/inflammation.zip
Once done, you can unzip this file using the
unzip command in Bash, which will unpack all the files
in this zip archive into the current directory:unzip inflammation.zip
This will unpack the zip file's contents into the new
inflammation directory. The file structure should look like this:inflammation/ ├── data │ ├── inflammation-*.csv ├── inflammation │ ├── __init__.py │ ├── models.py │ └── views.py ├── tests │ ├── __init__.py │ ├── test_patient.py │ └── test_models.py ├── .gitignore ├── LICENSE ├── README.md └── requirements.txt
The only files we'll be working with in this course are the following, so you can ignore the rest for now:
inflammation/models.py- contains the functions we'll be testingtests/test_models.py- contains the tests we'll be writingdata/inflammation-*.csv- contains the data we'll be using to test our functions
What Does the Patient Inflammation Data Contain?
Each dataset records inflammation measurements from a separate clinical trial of the drug, and each dataset contains information for 60 patients, who had their inflammation levels recorded for 40 days whilst participating in the trial.
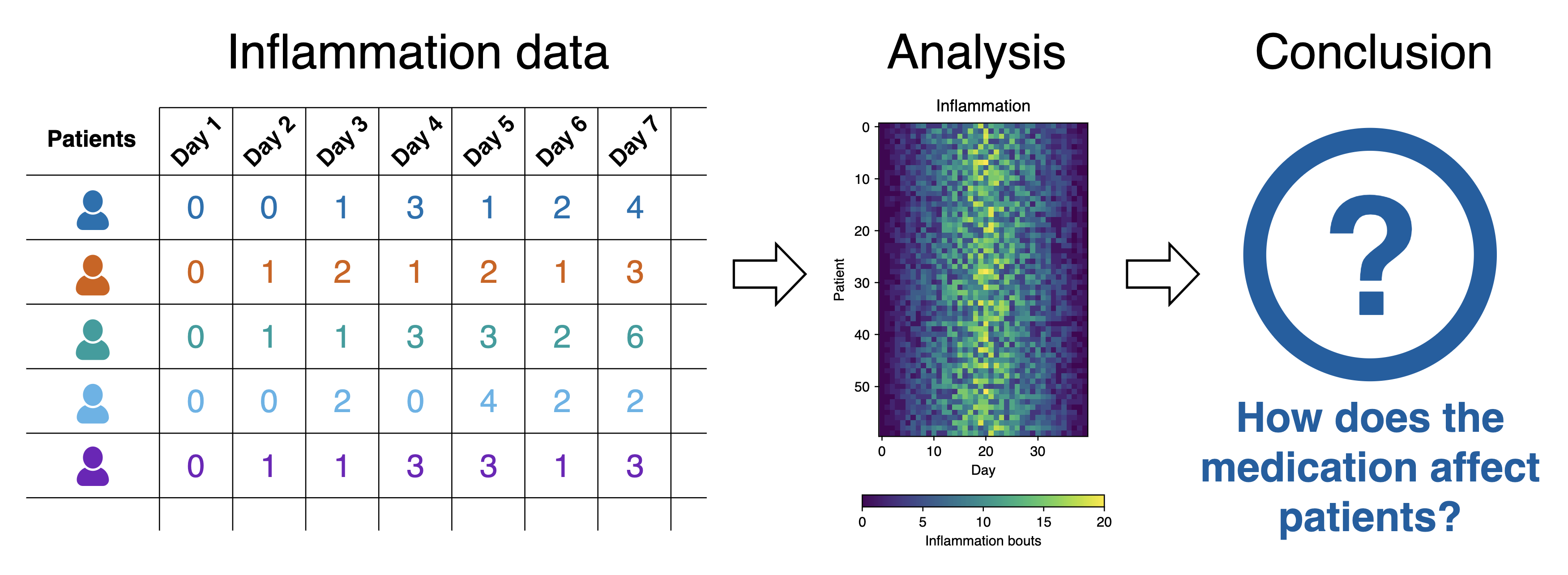 Inflammation study pipeline from the Software Carpentry Python novice lesson
Inflammation study pipeline from the Software Carpentry Python novice lessonEach of the data files uses the popular comma-separated (CSV) format to represent the data, where:
- Each row holds inflammation measurements for a single patient,
- Each column represents a successive day in the trial,
- Each cell represents an inflammation reading on a given day for a patient (the number of bouts of inflammation on that day).
The data is based on a clinical trial of inflammation in patients who have
been given a new treatment for arthritis. There are a number of datasets in the
data directory recording inflammation information in patients (each file
representing a different trial), and are each stored in comma-separated values
(CSV) format: each row holds information for a single patient, and the columns
represent successive days when inflammation was measured in patients.Let's take a quick look at the data now from within the Python command line
console. Change directory to your project, create a new virtual environment and
activate it. Install the required dependencies (
numpy and matplotlib) and
then start the Python console by invoking the Python interpreter without any
parameters, e.g.:python3 -m venv venv source venv/bin/activate pip install numpy matplotlib python
The last command will start the Python console within your shell, which enables us to execute Python commands
interactively. Inside the console enter the following:
import numpy as np data = np.loadtxt(fname='data/inflammation-01.csv', delimiter=',') data.shape
(60, 40)
The data in this case is two-dimensional - it has 60 rows (one for each patient)
and 40 columns (one for each day). Each cell in the data represents an
inflammation reading on a given day for a patient.
Our patient inflammation application has a number of statistical functions held
in
inflammation/models.py: daily_mean(), daily_max() and daily_min(),
for calculating the mean average, the maximum, and the minimum values for a
given number of rows in our data. For example, the daily_mean() function looks
like this:def daily_mean(data): """Calculate the daily mean of a 2D inflammation data array for each day. :param data: A 2D data array with inflammation data (each row contains measurements for a single patient across all days). :returns: An array of mean values of measurements for each day. """ return np.mean(data, axis=0)
Here, we use NumPy's
np.mean() function to calculate the mean vertically
across the data (denoted by axis=0), which is then returned from the function.
So, if data was a NumPy array of three rows like...[[1, 2], [3, 4], [5, 6]]
...the function would return a 1D NumPy array of
[3, 4] - each value representing the mean of each column (which are, coincidentally, the same values as the second row in the above data array).To show this working with our patient data, we can use the function like this, passing the first four patient rows to the
function in the Python console:
from inflammation.models import daily_mean daily_mean(data[0:4])
Note we use a different form of
import here - only importing the daily_mean
function from our models instead of everything. This also has the effect that
we can refer to the function using only its name, without needing to include the
module name too (i.e. inflammation.models.daily_mean()).The above code will return the mean inflammation for each day column across the
first four patients (as a 1D NumPy array of shape (40, 0)):
array([ 0. , 0.5 , 1.5 , 1.75, 2.5 , 1.75, 3.75, 3. , 5.25, 6.25, 7. , 7. , 7. , 8. , 5.75, 7.75, 8.5 , 11. , 9.75, 10.25, 15. , 8.75, 9.75, 10. , 8. , 10.25, 8. , 5.5 , 8. , 6. , 5. , 4.75, 4.75, 4. , 3.25, 4. , 1.75, 2.25, 0.75, 0.75])
The other statistical functions are similar. Note that in real situations
functions we write are often likely to be more complicated than these, but
simplicity here allows us to reason about what's happening - and what we need to
test - more easily.
Let's now look into how we can test each of our application's statistical
functions to ensure they are functioning correctly.
Writing Tests to Verify Correct Behaviour
One Way to Do It?
One way to test our functions would be to write a series of checks or tests,
each executing a function we want to test with known inputs against known valid
results, and throw an error if we encounter a result that is incorrect. So,
referring back to our simple
daily_mean() example above, we could use [[1, 2], [3, 4], [5, 6]] as an input to that function and check whether the result
equals [3, 4]:import numpy.testing as npt test_input = np.array([[1, 2], [3, 4], [5, 6]]) test_result = np.array([3, 4]) npt.assert_array_equal(daily_mean(test_input), test_result)
So we use the
assert_array_equal() function - part of NumPy's testing library - to test that our calculated result is the same as our expected result. This
function explicitly checks the array's shape and elements are the same, and
throws an AssertionError if they are not. In particular, note that we can't
just use == or other Python equality methods, since these won't work properly
with NumPy arrays in all cases.We could then add to this with other tests that use and test against other
values, and end up with something like:
test_input = np.array([[2, 0], [4, 0]]) test_result = np.array([2, 0]) npt.assert_array_equal(daily_mean(test_input), test_result) test_input = np.array([[0, 0], [0, 0], [0, 0]]) test_result = np.array([0, 0]) npt.assert_array_equal(daily_mean(test_input), test_result) test_input = np.array([[1, 2], [3, 4], [5, 6]]) test_result = np.array([3, 4]) npt.assert_array_equal(daily_mean(test_input), test_result)
However, if we were to enter these in this order, we'll find we get the following after the first test:
... AssertionError: Arrays are not equal Mismatched elements: 1 / 2 (50%) Max absolute difference: 1. Max relative difference: 0.5 x: array([3., 0.]) y: array([2, 0])
This tells us that one element between our generated and expected arrays doesn't
match, and shows us the different arrays.
We could put these tests in a separate script to automate the running of these
tests. But a Python script halts at the first failed assertion, so the second
and third tests aren't run at all. It would be more helpful if we could get data
from all of our tests every time they're run, since the more information we
have, the faster we're likely to be able to track down bugs. It would also be
helpful to have some kind of summary report: if our set of tests - known as a
test suite - includes thirty or forty tests (as it well might for a complex
function or library that's widely used), we'd like to know how many passed or
failed.
Going back to our failed first test, what was the issue? As it turns out, the
test itself was incorrect, and should have read:
test_input = np.array([[2, 0], [4, 0]]) test_result = np.array([3, 0]) npt.assert_array_equal(daily_mean(test_input), test_result)
Which highlights an important point: as well as making sure our code is
returning correct answers, we also need to ensure the tests themselves are also
correct. Otherwise, we may go on to fix our code only to return an incorrect
result that appears to be correct. So a good rule is to make tests simple
enough to understand so we can reason about both the correctness of our tests as
well as our code. Otherwise, our tests hold little value.
Using a Testing Framework
Keeping these things in mind, here's a different approach that builds on the
ideas we've seen so far but uses a unit testing framework. In such a
framework we define our tests we want to run as functions, and the framework
automatically runs each of these functions in turn, summarising the outputs. And
unlike our previous approach, it will run every test regardless of any
encountered test failures.
Most people don't enjoy writing tests, so if we want them to actually do it, it must be easy to:
- Add or change tests,
- Understand the tests that have already been written,
- Run those tests, and
- Understand those tests' results
Test results must also be reliable. If a testing tool says that code is working
when it's not, or reports problems when there actually aren't any, people will
lose faith in it and stop using it.
Look at
tests/test_models.py:"""Tests for statistics functions within the Model layer.""" import numpy as np import numpy.testing as npt def test_daily_mean_zeros(): """Test that mean function works for an array of zeros.""" from inflammation.models import daily_mean test_input = np.array([[0, 0], [0, 0], [0, 0]]) test_result = np.array([0, 0]) # Need to use NumPy testing functions to compare arrays npt.assert_array_equal(daily_mean(test_input), test_result) def test_daily_mean_integers(): """Test that mean function works for an array of positive integers.""" from inflammation.models import daily_mean test_input = np.array([[1, 2], [3, 4], [5, 6]]) test_result = np.array([3, 4]) # Need to use NumPy testing functions to compare arrays npt.assert_array_equal(daily_mean(test_input), test_result) ...
Here, although we have specified two of our previous manual tests as separate
functions, they run the same assertions. Each of these test functions, in a
general sense, are called test cases - these are a specification of:
- Inputs, e.g. the
test_inputNumPy array - Execution conditions - what we need to do to set up the testing environment to run our test, e.g. importing the
daily_mean()function so we can use it. Note that for clarity of testing environment, we only import the necessary library function we want to test within each test function - Testing procedure, e.g. running
daily_mean()with ourtest_inputarray and usingassert_array_equal()to test its validity - Expected outputs, e.g. our
test_resultNumPy array that we test against
Also, we're defining each of these things for a test case we can run
independently that requires no manual intervention.
Going back to our list of requirements, how easy is it to run these tests? We
can do this using a Python package called
pytest. Pytest is a testing
framework that allows you to write test cases using Python. You can use it to
test things like Python functions, database operations, or even things like
service APIs - essentially anything that has inputs and expected outputs. We'll
be using Pytest to write unit tests, but what you learn can scale to more
complex functional testing for applications or libraries.What About Unit Testing in Other Languages?
Other unit testing frameworks exist for Python, including Nose2 and Unittest,
and the approach to unit testing can be translated to other languages as well,
e.g. FRUIT for Fortran, JUnit for Java (the original unit testing framework),
Catch for C++, etc.
Installing Pytest
You can install
pytest using pip - exit the Python console first (either with Ctrl-D or by typing exit()), then do:pip install pytest
Whether we do this via VsCode or the command line, the results are exactly the same: our virtual environment will now have the
pytest package installed for use.Running Tests
Now we can run these tests using
pytest:python -m pytest tests/test_models.py
Here, we use
-m to invoke the pytest installed module, and specify the tests/test_models.py file to run the tests in that file
explicitly.Why Run Pytest Using python -m and Not pytest ?
Another way to run
pytest is via its own command, so we could try to use pytest tests/test_models.py on the
command line instead, but this would lead to a ModuleNotFoundError: No module named 'inflammation'. This is because
using the python -m pytest method adds the current directory to its list of directories to search for modules,
whilst using pytest does not - the inflammation subdirectory's contents are not 'seen', hence the
ModuleNotFoundError. There are ways to get around this with various methods, but we've used python -m for simplicity.============================================== test session starts ===================================================== platform darwin -- Python 3.9.6, pytest-6.2.5, py-1.11.0, pluggy-1.0.0 rootdir: /Users/alex/python-intermediate-inflammation plugins: anyio-3.3.4 collected 2 items tests/test_models.py .. [100%] =============================================== 2 passed in 0.79s ======================================================
Pytest looks for functions whose names also start with the letters 'test_' and
runs each one. Notice the
.. after our test script:- If the function completes without an assertion being triggered, we count the test as a success (indicated as
.). - If an assertion fails, or we encounter an error, we count the test as a failure (indicated as
F). The error is included in the output so we can see what went wrong.
So if we have many tests, we essentially get a report indicating which tests
succeeded or failed. Going back to our list of requirements, do we think these
results are easy to understand?
Write Some Unit Tests
We already have a couple of test cases in
test/test_models.py that test the
daily_mean() function. Looking at inflammation/models.py, write at least two
new test cases that test the daily_max() and daily_min() functions, adding
them to test/test_models.py. Here are some hints:- You could choose to format your functions very similarly to
daily_mean(), defining test input and expected result arrays followed by the equality assertion. - Try to choose cases that are suitably different, and remember that these functions take a 2D array and return a 1D array with each element the result of analysing each column of the data.
Once added, run all the tests again with
python -m pytest tests/test_models.py, and you should also see your new tests pass.The big advantage is that as our code develops we can update our test cases and
commit them back, ensuring that ourselves (and others) always have a set of
tests to verify our code at each step of development. This way, when we
implement a new feature, we can check a) that the feature works using a test we
write for it, and b) that the development of the new feature doesn't break any
existing functionality.
What About Testing for Errors?
There are some cases where seeing an error is actually the correct behaviour,
and Python allows us to test for exceptions. Add this test in
tests/test_models.py:import pytest ... def test_daily_min_string(): """Test for TypeError when passing strings""" from inflammation.models import daily_min with pytest.raises(TypeError): error_expected = daily_min([['Hello', 'there'], ['General', 'Kenobi']])
Note that you need to import the
pytest library at the top of our
test_models.py file with import pytest so that we can use pytest's
raises() function.Run all your tests as before.
Why Should We Test Invalid Input Data?
Testing the behaviour of inputs, both valid and invalid, is a really good idea
and is known as data validation. Even if you are developing command line
software that cannot be exploited by malicious data entry, testing behaviour
against invalid inputs prevents generation of erroneous results that could lead
to serious misinterpretation (as well as saving time and compute cycles which
may be expensive for longer-running applications). It is generally best not to
assume your user's inputs will always be rational.
Key Points
- The three main types of automated tests are unit tests, functional tests and regression tests.
- We can write unit tests to verify that functions generate expected output given a set of specific inputs.
- It should be easy to add or change tests, understand and run them, and understand their results.
- We can use a unit testing framework like Pytest to structure and simplify the writing of tests in Python.
- We should test for expected errors in our code.
- Testing program behaviour against both valid and invalid inputs is important and is known as data validation.