ODE Solvers - PM Exercises
In these exercises we will use Python to solve a variety of Ordinary Differential
Equations (ODEs), including initial and boundary value problems. Prior to attempting the
exercises you should be familiar with how to solve first order ODEs using separable
solutions and integrating factors. You should also know how solve a second order ODE
with constant coefficients and how to reduce a general second order ODE to a system of
coupled first order equations. If you are not familiar with this material then speak to
a module leader or demonstrator who will give you a tutorial. Advanced exercises are
marked with a star (); you should attempt these only if you have time and have
completed the other exercises.
Motivation
Ordinary differential equations are used very frequently in modelling biological
and physiological processes. Most commonly they are used to model the way in
which quantities of interest (such as concentrations of drugs, viral load, or
population densities) change as a function of time. The earlier exercises below
are revision of the kinds of ODEs you may have encountered at A level or as an
undergraduate. The later exercises are taken from real models of chemical and
biological systems.
The intentions behind this exercise are:
- To give you a clear goal to meet
- To give you a chance to revise or learn a few things. Specifically:
- The abstract base class pattern;
- Unit testing with
unittest; - Ordinary differential equation solver implementations.
A nice feature of this assignment is that you are provided with a testing
infrastructure which gives an
implicit specification of the code you are required to write. This means that
when you've written code
such that all the tests pass then you will know that you have completed the
exercise.
Initial value problems
Solve the following initial value problems by hand:
- , subject to .
- , subject to .
- , subject to .
- and , subject to and .
- , subject to and .
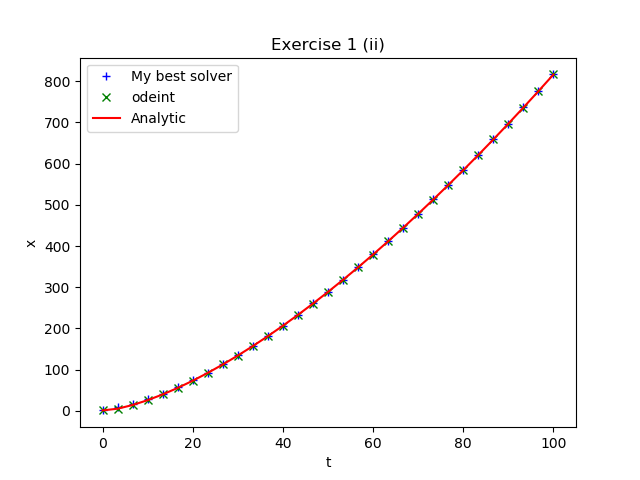
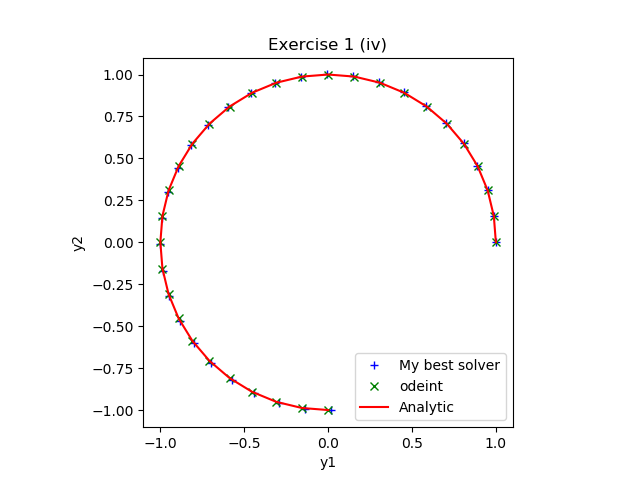
Solve each of the above initial value problems numerically using
your best Python ODE solver and compare with the analytic
solutions. Note that in order to solve the last problem numerically
you will need to reformulate the equation as a system of first order
ODEs. See if, for some of your answers, you can make a figure similar to those below,
made using model solutions.
Boundary value problems
Use
scipy.integrate.solve_bvp to solve the boundary value problemsubject to the boundary conditions and .
Solve the same boundary value problem, but now with the boundary conditions
and .
Chemical reaction systems
Mathematical models of simple chemical or biochemical reaction mechanisms often take the
form of non-linear systems of ordinary differential equations (derived using the
standard chemical laws of mass action). Often the various reactions making up the system
happen on very different time scales leading to a stiff system. An example is
Robertson's chemical reaction model, in which the concentrations of three reacting
chemical species evolve according to the system of equations
with initial conditions
Note: your mileage may vary with this question, because with more modern versions
of
scipy it becomes increasingly hard to stop the integration
scheme being "clever" and using a sophisticated scheme earlier.- Read the documentation for
scipy.integrate.ode. Solve this system using the Dormand & Princedoprisolver, which is a high-order Runge-Kutta solver, until in steps of . (Note that the interface is a bit more difficult to set up thanodeintbut an example is given with the documentation.) What warning do you get from the solver? Can you change any of thedopriparameters to get rid of warnings? How long does the integrator take to solve the system? - If you are still getting warnings for the
doprithen you will have a hint to help you select a betterscipy.integrate.odemethod. Switch to such a method. Again work to remove all warnings. - () Explain what is happening mathematically and chemically.
- Repeat parts (a)--(b) for the systemwith the same initial conditions. Which solver is faster now?
Which solver gives you warnings now? - Which solver should you use in which situation?
- () If you are feeling brave then assess how one of the fixed time-step solvers you have written yourself measures up to the solver you used in part (b). There are 3 species so you may want to extend the functionality to cope with . Use the solution from part (b) as a reference to measure your error. How small do you need to make your time-step to get within a particular error?
When Zombies Attack!
()Inspired by a famous SIR model for epidemics there is a
SZR model for
zombie invasion. The paper describing this model is
available to download from
https://mysite.science.uottawa.ca/rsmith43/Zombies.pdf
which includes modelling code in
Matlab.A more advanced model included in the paper, which includes latent infection and
quarantine, is known as the SIZRQ model. This model is defined by
with initial conditions
and parameter values
- Solve this system of equations numerically.
- How realistic is this type of model? Can you think of any improvements to the model?
Simple events
()ODE-based models of biological systems often include specific events,
which may occur either at predetermined times or when a given
condition on the values of the state variables or their derivatives is
met. The following model describes a zombie outbreak in which there is
periodic culling (Spoiler alert. It looks like we all die
whatever we do.
If you think that's depressing then you might have to read this
followup paper).
of the zombie population:
Here is the kill ratio, and the last equation represents periodic culling of the zombie population.
Solve this model subject to initial conditions
and with parameter values
and culling every 10 units of time (i.e. , , etc.). Hint: you will need to loop over time intervals to solve this model.